Multiwfn forum
Multiwfn official website: http://sobereva.com/multiwfn. Multiwfn forum in Chinese: http://bbs.keinsci.com/wfn
You are not logged in.
- Topics: Active | Unanswered
#1 2020-06-24 04:16:09
- dweera87
- Member
- Registered: 2020-06-24
- Posts: 5
Electron density difference projected on to a line
Dear Multiwfn Developers,
I have been following section 4.5.5 in the manual to create electron density difference maps of heterojuctions of graphitic carbon nitride and phosphorene quantum dots. (Δρ(r)= ρ_tot(r)- ρ_phossphorene(r)- ρ_graphitc carbon nitride(r))
I was trying to project electron density difference of such a system on to a perpendicular line. For example if my 2D quantum dots lie in the xy plane, I want to project the electron density difference of the system to a line parallel to the z axis.
(Figure 5c of RSC Adv., 2020,10,5260-5267)
I was trying a similar procedure as in section 4.5.5 in the manual but used option 3 in the main menu (plot in a line) rather than 5(calculate grid data). However, I noticed that my plots vary strangely depending on the x,y,z coordinates of the two points that define the line.
Could you please let me know if this procedure would create reliable results for my problem or if this method is fundamentally wrong to apply for my problem?
Thank you in advance!
Best Regards,
Dimuthu
Offline
#2 2020-06-24 07:25:55
Re: Electron density difference projected on to a line
Dear Dimuthu,
The Figure 5c of the RSC Adv. paper cannot be plotted by main function 3. You should generate grid data of electron density difference first using main function 5, and then use subfunction 18 of main function 13 to convert the grid data to local integral curve. Please check Section 3.16.14 of Multiwfn manual for introduction and Section 4.13.6 for application example.
Best,
Tian
Offline
#3 2020-06-26 04:26:48
- dweera87
- Member
- Registered: 2020-06-24
- Posts: 5
Re: Electron density difference projected on to a line
Dear Dr. Tian Lu,
Thank you so much for your directions. I did go through sections 3.16.14 of Multiwfn manual for introduction and Section 4.13.6 for application example. I just noticed that my graphitic carbon nitride and phosphorene quantum dots do not lie exactly in the xy plane and therefore a perpendicular line drawn to the interface does not align with the z-axis. Is there an option in Multiwfn to transform a molecular system in to the desired orientation?
Thank you! Highly appreciate your advice!
Best,
Dimuthu
Offline
#4 2020-06-26 11:42:41
Re: Electron density difference projected on to a line
Dear Dimuthu,
You can use my VMD script to reorientate your system, so that the graphitic carbon nitride surface is parallel to XY plane.
Download VMD program (http://www.ks.uiuc.edu/Research/vmd/), boot up it and copy below commands into VMD console window to define a new command named "alignplane"
proc alignplane {ind1 ind2 ind3} {
set atm1 [atomselect top "serial $ind1"]
set atm2 [atomselect top "serial $ind2"]
set atm3 [atomselect top "serial $ind3"]
set vec1x [expr [$atm2 get x] - [$atm1 get x]]
set vec1y [expr [$atm2 get y] - [$atm1 get y]]
set vec1z [expr [$atm2 get z] - [$atm1 get z]]
set vec2x [expr [$atm3 get x] - [$atm1 get x]]
set vec2y [expr [$atm3 get y] - [$atm1 get y]]
set vec2z [expr [$atm3 get z] - [$atm1 get z]]
set sel [atomselect top all]
$sel move [transvecinv [veccross "$vec1x $vec1y $vec1z" "$vec2x $vec2y $vec2z"]]
$sel move [transaxis y 90]
} Load structure file of your system (.pdb/.xyz/.mol2, etc.) into VMD, assume that 3 10 12 are three atoms in the graphitic carbon nitride, run this command in VMD console window:
alignplane 3 10 12Now you can find the graphitic carbon nitride has been exactly parallel to XY plane. Choose "file" - "save Coordinate" to save to the present structure to a new geometry file.
Then, use your quantum chemistry code to perform single point task for this structure (as well as its fragments or other states) to obtain corresponding wavefunction files, then you will be able to use Multiwfn to plot local integral curve along the direction perpendicular to the surface.
Note that if you use Gaussian to perform the calculation, do not forget to add "nosymm" keyword to avoid automatic reorientation of the system to standard orientation.
Best,
Tian
Offline
#5 2020-11-06 17:01:44
- dweera87
- Member
- Registered: 2020-06-24
- Posts: 5
Re: Electron density difference projected on to a line
Dear Dr. Tian Lu,
Thank you so much for the VMD code that transforms my system into xy plane. I attached a charge displacement curve I obtained for one of my systems. (P20H12 + melamine) The CDC values are very large and I was just wondering if that could be right.
Could you please let me know if there is an obvious mistake that I have made?
How can I get an idea about the amount of charge transferred in the system?
Thank you!
Best,
Dimuthu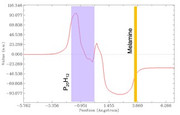
Offline
#6 2020-11-06 18:09:44
Re: Electron density difference projected on to a line
Dear Dimuthu,
I think the grid data of electron density difference you generated is problematic. After calculating the grid data, please do not forget to visualize its isosurface to make sure it is correct, you can also upload the isosurface map so that I can check.
Best,
Tian
Offline
#7 2020-11-06 18:32:05
- dweera87
- Member
- Registered: 2020-06-24
- Posts: 5
Re: Electron density difference projected on to a line
Dear Dr. Tian Lu,
I uploaded the isosurface map here. To create this, I input the P20H12-melamine.fchk file, choose 5 from the menu to calculate grid data, then choose 0 and substract the P20H12.fchk and melamine.fchk files and then choose 1 to calculate electron density.
Please let me know about any mistakes I'm making in the process.
Thank you!
Best,
Dimuthu
Offline
#8 2020-11-07 06:42:17
Re: Electron density difference projected on to a line
Your grid data of electron density difference is evidently wrong. You should guarantee that the atomic coordinates in the whole system and in the two monomers completely consistent with each other, however currently this requirement is violated. In order to properly calculate electron density difference, you should optimize the whole system first, then manually extract the atomic coordinates of the two monomers. Then, in the single point calculation of the two monomers, if you are using Gaussian, you should add "nosymm" keyword, otherwise Gaussian will automatically translate and reorientate the system, leading to discrepancy between the dimer and monomer coordinates.
Please carefully check the example in Section 4.5.5 of manual, a complete example of generating grid data of electron density difference is given.
Offline
#9 2020-11-09 16:18:21
- dweera87
- Member
- Registered: 2020-06-24
- Posts: 5
Re: Electron density difference projected on to a line
Dr. Tian Lu,
Thanks so much for pointing out what I was doing wrong. I'll check the Section 4.5.5.
Best,
Dimuthu
Offline