Multiwfn forum
Multiwfn official website: http://sobereva.com/multiwfn. Multiwfn forum in Chinese: http://bbs.keinsci.com/wfn
You are not logged in.
- Topics: Active | Unanswered
#1 2021-05-05 15:04:09
- Hans
- Member
- Registered: 2018-10-24
- Posts: 10
How to run a RELAXED SCAN in Gaussian 09?
I want to perform a RELAXED SCAN calculation between 12-crown-4 and lithium cation in Gaussian 9 consisting of the following:
As a starting point, placing the Li+ at a distance of 0.4 Å from the center of 12-crown-4 to a distance of 5.4 Å from said center as the end point of the calculation, with 0.4 Å increments (Fig. 1).
In other words, I want to perform the relaxed scan calculation by placing the Li+ at a slightly different distance from the center of the 12-crown-4 (this for symmetry reasons, to avoid a reorientation of the molecular system in the course of the calculation) up to a distance farthest from the ring. The problem I have is that I don't know how to type or what keywords should I use in Gaussian 9's .gjf file to run this calculation.
I attach the optimized geometry at the hf/3-21g level in salt.gjf (it is D2d symmetry).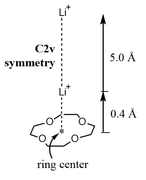
%chk=C:\salt.chk
# hf/3-21g
Title Card Required
1 1
Li 0.00000000 0.00000000 0.00000000
O 1.80378214 -0.00000000 0.32927408
C 2.30761918 1.26176110 -0.19253296
C 2.30761918 -1.26176110 -0.19253296
C 1.26176110 2.30761918 0.19253296
C 1.26176110 -2.30761918 0.19253296
O 0.00000000 1.80378214 -0.32927408
O -0.00000000 -1.80378214 -0.32927408
C -1.26176110 2.30761918 0.19253296
C -1.26176110 -2.30761918 0.19253296
C -2.30761918 1.26176110 -0.19253296
C -2.30761918 -1.26176110 -0.19253296
O -1.80378214 -0.00000000 0.32927408
H 3.26664225 1.49572411 0.24426407
H 2.39390318 1.19789109 -1.26923404
H 3.26664225 -1.49572411 0.24426407
H 2.39390318 -1.19789109 -1.26923404
H 1.49572411 3.26664225 -0.24426407
H 1.19789109 2.39390318 1.26923404
H 1.49572411 -3.26664225 -0.24426407
H 1.19789109 -2.39390318 1.26923404
H -1.49572411 3.26664225 -0.24426407
H -1.19789109 2.39390318 1.26923404
H -1.49572411 -3.26664225 -0.24426407
H -1.19789109 -2.39390318 1.26923404
H -3.26664225 1.49572411 0.24426407
H -2.39390318 1.19789109 -1.26923404
H -3.26664225 -1.49572411 0.24426407
H -2.39390318 -1.19789109 -1.26923404
I also asked this question in researchgate but I can't find an answer:
Offline
#2 2021-05-05 15:56:47
Re: How to run a RELAXED SCAN in Gaussian 09?
This cannot be realized via G09, but can be realized by means of GIC feature of G16. The input file should be
# B3LYP/6-31G* opt=addgic
Title Card Required
1 1
[coordinate part]
XCR(inactive)=XCntr(2-25)
YCR(inactive)=YCntr(2-25)
ZCR(inactive)=ZCntr(2-25)
scan(StepSize=0.4,NSteps=12)=sqrt[(X(1)-XCR)^2+(Y(1)-YCR)^2+(Z(1)-ZCR)^2]*0.529177where Li is atom 1, and 12-crown-4 is assumed to be atoms 2~25. The relaxed scan will be performed between Li and center of 12-crown-4, the initial distance corresponds to the inputted coordinate, the stepsize is 0.4 Angstrom, and number of steps is 12.
If you can only use G09, unfortunately it is impossible to realize the relaxed scan as you expected, you can only perform rigid scan, namely scanning Z coordinate of Li while keep positions of atoms of 12-crown-4 fixed.
Offline