Multiwfn forum
Multiwfn official website: http://sobereva.com/multiwfn. Multiwfn forum in Chinese: http://bbs.keinsci.com/wfn
You are not logged in.
- Topics: Active | Unanswered
#1 Re: Multiwfn and wavefunction analysis » Viewing Molecular Orbitals diagram » 2023-02-05 18:45:03
Dear Professor
Thank you for reply.
Best Regards
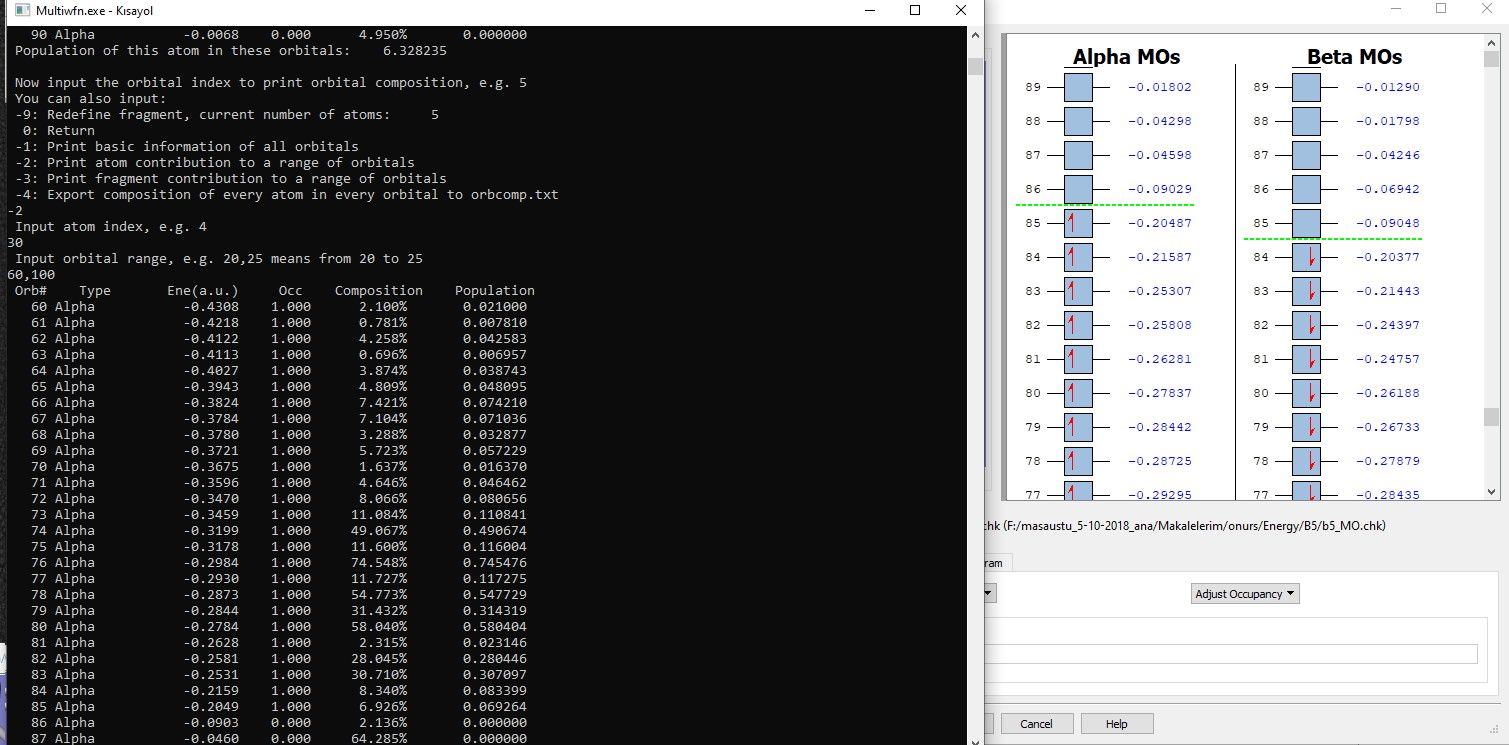
#2 Multiwfn and wavefunction analysis » Viewing Molecular Orbitals diagram » 2023-02-05 13:29:51
- taha55
- Replies: 2
#3 Re: Multiwfn and wavefunction analysis » Fukui function » 2022-12-01 13:29:47
Dear Professor
Thank you very much
#4 Multiwfn and wavefunction analysis » Fukui function » 2022-12-01 11:05:49
- taha55
- Replies: 2
Dear Professor
When calculating fukui(as section 4.22.1 in manuel) , in the current version(3.8), there are no 7, 10 and 16 options after the 100 command as below. Can you fix it?
.
.
.
.
Boot up Multiwfn and input below commands:
examples\phenol.xyz
100 // Other functions (Part 1)
16 // Calculate various quantities in conceptual density functional theory
1 // Generate .wfn files for N, N+1, N-1 electrons states
.
.
.
Best Regards
#5 Re: Quantum Chemistry » cp2k output trajectory convert to pdb » 2022-09-30 11:39:49
Thank you Dear Prefessor
Best Regards
#6 Re: Quantum Chemistry » cp2k output trajectory convert to pdb » 2022-09-29 20:36:53
Thank you Dear Prefessor
I know what you said, but it takes hours to open vmd xyz file at 100000 frames. How to do it differently from VMD?
#7 Quantum Chemistry » cp2k output trajectory convert to pdb » 2022-09-29 19:54:18
- taha55
- Replies: 4
Hi
How to convert output trajectory(Molecular_dynamic.xyz file) to pdb format?
Regards
#8 Re: Multiwfn and wavefunction analysis » QTAIM topology analysis for excited state calculated by CP2K » 2022-06-12 22:19:26
Dear Professor
Thank you very much for your help and update
Best Regards
#9 Re: Multiwfn and wavefunction analysis » QTAIM topology analysis for excited state calculated by CP2K » 2022-06-11 13:54:26
Can you help me on how to do this? how to get lattice vectors?
α=γ=90°, β=103°
a=21 b=11 and c=19 angstrom
#10 Re: Multiwfn and wavefunction analysis » QTAIM topology analysis for excited state calculated by CP2K » 2022-06-11 11:31:55
Dear Professor
my file
[Molden Format]
[Cell]
13.0500000 0.00000000 0.00000000
0.00000000 13.0500000 0.00000000
0.00000000 0.00000000 13.0500000
The unit cell lengths of the cubic structure were entered as above. But if the structure is not cubic, How to enter the angle values of the unit cell for a monoclinic structure(α=γ=90°≠β)?
Best Regards
#11 Re: Multiwfn and wavefunction analysis » QTAIM topology analysis for excited state calculated by CP2K » 2022-03-27 12:35:20
Dear Professor
Thank you very much for your help and explanation
Best Regards
#12 Re: Multiwfn and wavefunction analysis » QTAIM topology analysis for excited state calculated by CP2K » 2022-03-27 11:18:09
Dear Professor;
You contribute a lot to science and scientists. Really sincerely thank you very much. I want to ask one last thing about this subject. My molden file has 2 atoms Oxygen and Hydrogen. Could you provide some information on what is the [Nval] specified in the multiwfn manual. Also, are the numbers next to the O and H atoms (6 for O and 1 for H) the number of valence electrons?And did I write these numbers correctly?
my file
[Molden Format]
[Cell]
13.0500000 0.00000000 0.00000000
0.00000000 13.0500000 0.00000000
0.00000000 0.00000000 13.0500000
[Nval]
O 6
H 1
[Atoms] AU
O 1 8 0.000000 0.000000 5.680649
H 2 1 -1.609816 0.155501 6.775287
H 3 1 -0.042463 1.710742 3.727409
.
.
.
#13 Re: Multiwfn and wavefunction analysis » QTAIM topology analysis for excited state calculated by CP2K » 2022-03-26 15:09:45
#14 Re: Multiwfn and wavefunction analysis » QTAIM topology analysis for excited state calculated by CP2K » 2022-03-26 11:48:22
Dear Professor
I calculated cp for vmd using the diamond_222.molden file and as in your video (https://www.youtube.com/watch?v=mgsnhvWH5SI), but the vmd opened molecule was not periodic. I chose the x, y and z coordinates, but the periodic molecules did not form.
Best regards
#15 Re: Multiwfn and wavefunction analysis » QTAIM topology analysis for excited state calculated by CP2K » 2022-03-25 18:30:47
Dear Professor
I will calculate the aim topology, that is, BCP (bond critic point), using the ''diamond_222.molden'' file as in Section 4.2. Is there any need to convert the "diamond_222.molden" file to the "NO_0001.mwfn'' file for this? so we can't use diamond_222.molden file for BCP (bond critic point) calculation as Section 4.2?
Best Regards
#16 Re: Multiwfn and wavefunction analysis » QTAIM topology analysis for excited state calculated by CP2K » 2022-03-25 08:28:28
Dear Professor, thank you for reply
''Next, we need to generate a wavefunction file containing natural orbitals of the excited state of interest. Boot up Multiwfn and input
diamond_222.molden
18 // Electron excitation analysis
13 // Generate natural orbitals of excited states
diamond_222.out
1 // The excited state of interest is assumed to be the first excited state
[Press ENTER button] // Use 0.25 Bohr of grid spacing to generate overlap matrix''
Is the cp obtained by this method valid for a periodic system or for an isolated molecule in the gas phase?
Best regards
#17 Re: Multiwfn and wavefunction analysis » QTAIM topology analysis for excited state calculated by CP2K » 2022-03-24 13:50:19
Dear Prefessor
Many thanks for this information. I can calculate an organic molecule with multiwfn. In line with the information you have given above, will I have periodically made a PBC calculation using the calculations in section 4.2 of the NO_0001.mwfn file that I obtained with cp2k (using PBC=periyodic boundry condidation)?
Best Regards
#18 Re: Multiwfn and wavefunction analysis » ESP map Fukui functions » 2020-04-06 09:23:37
Professsor Tian Lu, thank you very much
Best Regard
#19 Re: Multiwfn and wavefunction analysis » ESP map Fukui functions » 2020-04-05 11:46:40
Professsor Tian Lu, thank you very much
I ask this question independently of ESP. Many articles are described differently.
So,is the following conclusion correct for Fukui functions?
F+→ easily undergo nukleophilic attack (i.e. nukleophilic reagents tend to interact with this site)
F-→ easily undergo electrophilic attack (i.e. electrophilic reagents tend to interact with this site)
#20 Multiwfn and wavefunction analysis » ESP map Fukui functions » 2020-04-05 09:13:21
- taha55
- Replies: 4
Hello Professsor Tian Lu
Which of the following(1 or 2) is true that relationship between the red and blue regions of the MEP map and the fukui functions?
1. Red colour areas: electronically rich→nucleophile (proton-loving)→nucleophilic attack→f+ (fukui) values are positive and high
Blue colour areas: electronically poor→electrophile (electron-loving)→electrophilic attack→f- (fukui) values are positive and high
Or
2. Red colour areas: electronically rich→nucleophile (proton-loving)→electrophilic attack→f- (fukui) values are positive and high
Blue colour areas: electronically poor→electrophile (electron-loving)→nucleophilic attack→f+ (fukui) values are positive and high
Best Regards
#21 Re: Multiwfn and wavefunction analysis » multiwfn manual for reference » 2020-03-22 14:36:22
Thank you very much Professsor Tian Lu
#22 Re: Multiwfn and wavefunction analysis » multiwfn manual for reference » 2020-03-22 07:16:58
Thank you for reply Professsor Tian Lu. Do you have an article about ICSS and NICS calculating (section ''4.200.4 Study iso-chemical shielding surface (ICSS) and magnetic shielding distribution' in multiwfn manual')?
Best Regard
#23 Multiwfn and wavefunction analysis » multiwfn manual for reference » 2020-03-22 05:14:51
- taha55
- Replies: 4
Hello Professsor Tian Lu
I want to use some information in your multiwfn manual in my article. Can you suggest an your article that I will give reference my article?
Best Regard
#24 Re: Multiwfn and wavefunction analysis » VMD graphic representation » 2020-03-08 10:59:38
Dear Professor Tian Lu
After reviewing this video thoroughly, I succeeded.
Thank you for reply
Best regards
#25 Multiwfn and wavefunction analysis » VMD graphic representation » 2020-03-07 13:03:24
- taha55
- Replies: 2
#26 Re: Multiwfn and wavefunction analysis » Fukui f+, f- calculating formula » 2020-03-03 18:43:13
Thank you very much for help
Best Regards
#27 Re: Multiwfn and wavefunction analysis » Fukui f+, f- calculating formula » 2020-03-03 18:12:49
Thank you. Can you suggest an article or yours containing the correct formulas in Section 4.7.3 for me to cite?
Best Regards
#28 Multiwfn and wavefunction analysis » Fukui f+, f- calculating formula » 2020-03-03 08:45:16
- taha55
- Replies: 10
Hello Dear
The formulas for f+,f- which is many articles and multiwfn manual;
f+= q(N+1)− qN and f-= q(N)− q(N-1) (page 387 in multiwfn manual) (1. Eq.) (N=charge 0, N+1=charge-1, N-1= charge+1)
Fuki calculation is given below in section ''Calculating global and atomic indices ''(page 679) in multiwfn manual
Units used below are "e" (elementary charge)
Atom q(N) q(N+1) q(N-1) f- f+ f0 CDD
1(C ) -0.0587 -0.1185 0.0852 0.1439 0.0598 0.1018 -0.0841
2(C ) -0.0390 -0.1674 0.0268 0.0658 0.1284 0.0971 0.0626
3(C ) -0.0597 -0.1873 0.0319 0.0916 0.1276 0.1096 0.0360
If f+and f- are calculated for 1(C) according to equation 1;
f+= q(N+1)− qN=-0.1185-( -0.0587)=-0.0598
f-=q(N)− q(N-1)=-0.0587-0.0852=-0.1439
But obove the table are calculated as f+= 0.0598 and f-=0.1439 so Do these formulas must arranged as follows?
f+= qN(r)-q(N+1) (2. Eq.)
f- =q(N-1)-qN(r)
Which formula is correct for f+and f- (1. or 2. Eq.)?
Best regards
#29 Re: Multiwfn and wavefunction analysis » Beta spin review » 2020-02-25 11:09:12
Thank you very much for helping very helpful again
Best Wishes
#30 Multiwfn and wavefunction analysis » Beta spin review » 2020-02-24 20:56:06
- taha55
- Replies: 2